When you have a data frame with a column content grouped by a delimiter, you might want to split that into rows by using R. Not so frequent activity as splitting column into multiple columns by separator.
Here is my data frame. One of the columns contains a text grouped by the delimiter. I selected only those rows that are necessary for demonstration.
require(dplyr) mydf <- starwars %>% group_by(species) %>% summarise(all_names = paste(name, collapse = "; ")) %>% filter(row_number() %in% c(15:18))
I used the knitr package to create a beautiful output in the R console.
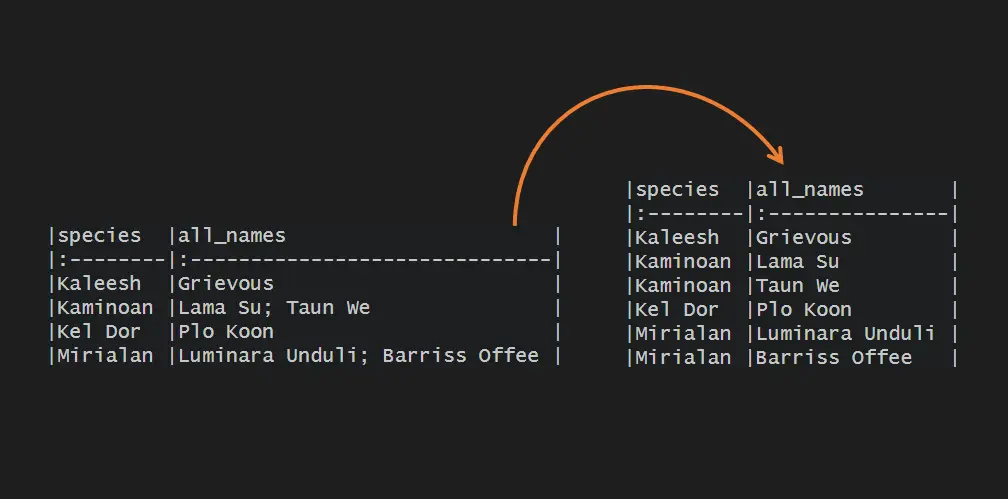
mydf %>% knitr::kable() #|species |all_names | #|:--------|:------------------------------| #|Kaleesh |Grievous | #|Kaminoan |Lama Su; Taun We | #|Kel Dor |Plo Koon | #|Mirialan |Luminara Unduli; Barriss Offee |
Column into rows by using R
The easiest way to separate a column into rows using R is using the seperate_rows function from the tidyr package. Be careful with the delimiter if there is also whitespace. Of course, you can remove that after splitting into rows, but why not at once.
mydf %>% tidyr::separate_rows(all_names, sep = "; ") ## A tibble: 6 x 2 # species all_names # #1 Kaleesh Grievous #2 Kaminoan Lama Su #3 Kaminoan Taun We #4 Kel Dor Plo Koon #5 Mirialan Luminara Unduli #6 Mirialan Barriss Offee
If you want to do the opposite of separate_rows in R, then look at this post.
Remember that a single backslash used as a delimiter is considered an escape character, and here is how to deal with that.
Split data frame by multiple columns into rows
What if you have multiple columns that you want to split into rows simultaneously?
mydf <- starwars %>%
group_by(species) %>%
summarise(
all_names = paste(name, collapse = "; "),
all_names2 = paste(name, collapse = ", ")
) %>%
filter(row_number() %in% c(15:18))
mydf %>% knitr::kable()
#|species |all_names |all_names2 |
#|:--------|:------------------------------|:------------------------------|
#|Kaleesh |Grievous |Grievous |
#|Kaminoan |Lama Su; Taun We |Lama Su, Taun We |
#|Kel Dor |Plo Koon |Plo Koon |
#|Mirialan |Luminara Unduli; Barriss Offee |Luminara Unduli, Barriss Offee |To do that, you can combine multiple seperate_rows functions, but as you can see, it creates all possible combinations.
mydf %>% tidyr::separate_rows(all_names, sep = "; ") %>% tidyr::separate_rows(all_names2, sep = ", ") ## A tibble: 10 x 3 # species all_names all_names2 # #1 Kaleesh Grievous Grievous #2 Kaminoan Lama Su Lama Su #3 Kaminoan Lama Su Taun We #4 Kaminoan Taun We Lama Su #5 Kaminoan Taun We Taun We #6 Kel Dor Plo Koon Plo Koon #7 Mirialan Luminara Unduli Luminara Unduli #8 Mirialan Luminara Unduli Barriss Offee #9 Mirialan Barriss Offee Luminara Unduli #10 Mirialan Barriss Offee Barriss Offee
To deal with that, you can use the distinct function from dplyr, but how to use it depends on the scenario. In my case, it looks like this.
mydf %>% tidyr::separate_rows(all_names, sep = "; ") %>% tidyr::separate_rows(all_names2, sep = ", ") %>% distinct(species, all_names,.keep_all = T) ## A tibble: 6 x 3 # species all_names all_names2 # #1 Kaleesh Grievous Grievous #2 Kaminoan Lama Su Lama Su #3 Kaminoan Taun We Lama Su #4 Kel Dor Plo Koon Plo Koon #5 Mirialan Luminara Unduli Luminara Unduli #6 Mirialan Barriss Offee Luminara Unduli
Take a look at this post if you want to do the opposite – concatenation by a group in R.
Leave a Reply