Sometimes it might be very confusing when it looks like R is ignoring the NA value. When some of the code relying on that values, the results might be confusing.
The problem, when it looks like R ignores the NA value, appears when some of the NA values are a string. It might happen after using some of the functions. If the column already contains NA, then sprintf might turn it into a string format.
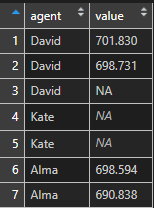
Here is my dataset that contains column values with missing values.
df <- data.frame(
agent = as.character(c("David", "David", "David", "Kate", "Kate", "Alma", "Alma")),
value = as.numeric(c(701.8299, 698.7306, NA, NA, 698.7125, 698.5938, 690.8382))
)
df
# agent value
#1 David 701.8299
#2 David 698.7306
#3 David NA
#4 Kate NA
#5 Kate 698.7125
#6 Alma 698.5938
#7 Alma 690.8382Here is an additional step with ifelse checking some logic. If it is not true, ifelse returns the column with improvements from sprintf.
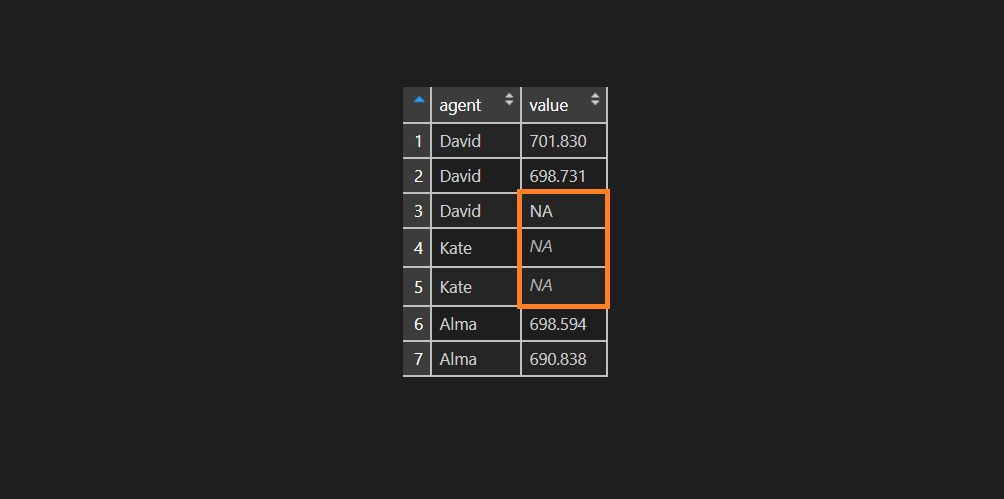
df$value <- ifelse(df$agent == "Kate", NA, sprintf("%.3f", df$value))As you can see, that visually there is some difference between NA values. There is no surprise that R ignoring NA value that only looks like NA.
is.na(df$value) FALSE FALSE FALSE TRUE TRUE FALSE FALSE
The fastest way to convert string NA values is with the function as.numeric.
df$value <- as.numeric(df$value)
NA values may cause some big headaches. Take a look at one of the examples with ifelse: ifelse and NA problem in R.
Leave a Reply