Here is a simple way how to combine CSV or text files with R and, at the same time, add a column with filenames. Here is all the code with more detailed explanations below.
require(dplyr)
require(data.table)
# read file path
all_paths <-
list.files(path = "~/txt_files/",
pattern = "*.txt",
full.names = TRUE)
# read file content
all_content <-
all_paths %>%
lapply(read.table,
header = TRUE,
sep = "\t",
encoding = "UTF-8")
# read file name
all_filenames <- all_paths %>%
basename() %>%
as.list()
# combine file content list and file name list
all_lists <- mapply(c, all_content, all_filenames, SIMPLIFY = FALSE)
# unlist all lists and change column name
all_result <- rbindlist(all_lists, fill = T)
# change column name
names(all_result)[3] <- "File.Path"I have 3 txt files, and each of them contains Tab-delimited movie data from IMDB.
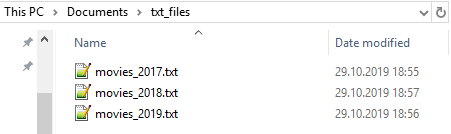
To combine files with R and add filename column, follow these steps.
1. Read paths to files
all_paths <-
list.files(path = "~/txt_files/",
pattern = "*.txt",
full.names = TRUE)2. Read file content
all_content <-
all_paths %>%
lapply(read.table,
header = TRUE,
sep = "\t",
encoding = "UTF-8")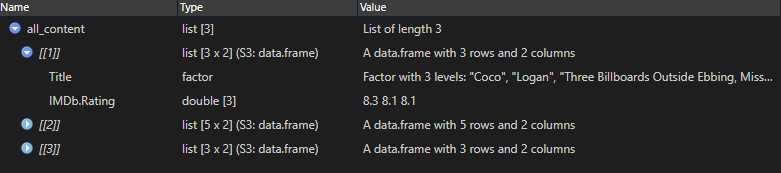
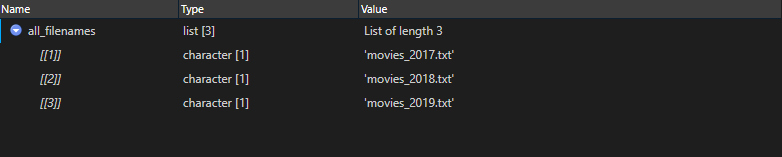
3. Read file names
all_filenames <- all_paths %>% basename() %>% as.list()
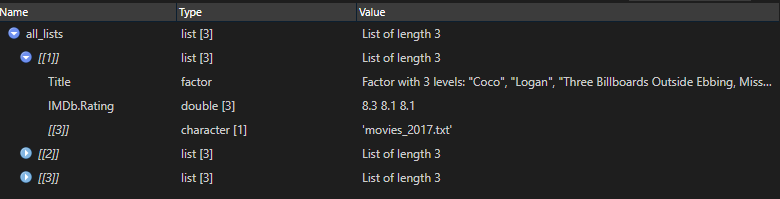
4. Combine file content list with filename list
all_lists <- mapply(c, all_content, all_filenames, SIMPLIFY = FALSE)
5. Unlist result and do some finalization
all_result <- rbindlist(all_lists, fill = T) # change column name names(all_result)[3] <- "File.Path"
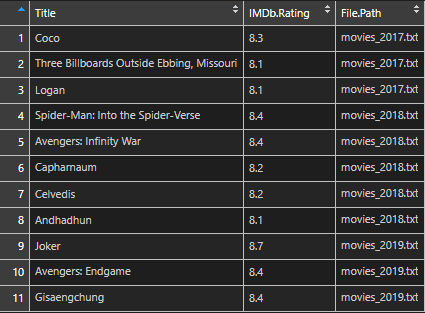
It is also possible to do that at once.
# all process in one
all_txt <- rbindlist(mapply(
c,
(
list.files(
path = "~/txt_files/",
pattern = "*.txt",
full.names = TRUE
) %>%
lapply(
read.table,
header = TRUE,
sep = "\t",
encoding = "UTF-8"
)
),
(
list.files(
path = "~/txt_files/",
pattern = "*.txt",
full.names = TRUE
) %>%
basename() %>%
as.list()
),
SIMPLIFY = FALSE
),
fill = T)Please, check other R related posts that might be interesting for you.
Leave a Reply